What we investigate
Our laboratory uses mass spectrometry-based proteomics to understand interconnected protease networks as signaling layers governing cell-cell communication in complex tissue responses. With a focus on inflammatory skin disorders and wound healing, we aim at devising new strategies for therapeutically intervening with detrimental protease activities, redefining their potential as drug targets.
KEYWORDS
degradomics, protease, proteomics, skin, wound healing
Our research in more detail
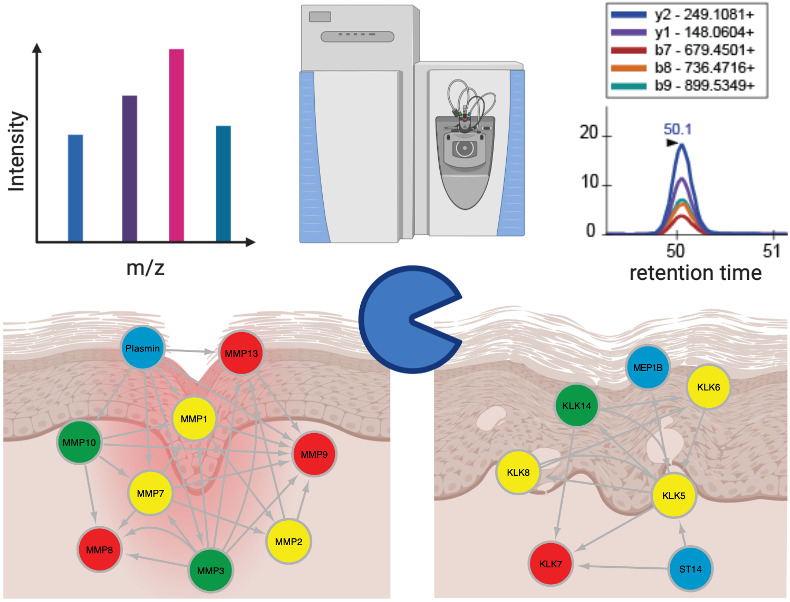
Proteases play pivotal roles in many diseases and form 5-10% of all potential drug targets. It is now clear that detrimental proteolysis results from perturbation of a complex proteolytic network rather than altered activity of a single enzyme. Disturbances of such networks lead to severe diseases including cancer and inflammatory disorders. However, so far technical limitations prohibited a global understanding of interconnected protease activity in complex systems, contributing to devastating failures of protease inhibitors in clinical trials.
Our research group develops and applies novel proteomics technologies to understand protease networks in inflamed tissues and hyperproliferative epithelia focusing on the skin as model system. Targeted assays discriminate pro- from active proteases, and our next-generation positional proteomics workflows allow analyzing small sample amounts from cells and tissues obtained from animal models and patient biopsies.
In translational research, we monitor and functionally characterize protease substrate cleavages as net outcomes of complex proteolytic activities in healing impairments, a common complication in elderly people and patients suffering from diabetes. In collaboration with clinicians, we apply approaches that we have implemented using clinically relevant wound models to the analysis of wound samples from patients with normal and delayed healing.

Prof. Ulrich auf dem Keller
Please contact SKINTEGRITY.CH coordinator
Dr. Maarten Schledorn with any questions
or inquiries.
Email
Selected publications
SKINTEGRITY.CH Principal Investigators are in bold:
- Mikosiński J, Kalogeropoulos K, Bundgaard L, Larsen CA, Savickas S, Moldt Haack A, Pańczak K, Rybołowicz K, Grzela T, Olszewski M, Ciszewski P, Sitek-Ziółkowska K, Twardowska-Saucha K, Karczewski M, Rabczenko D, Segiet A, Buczak-Kula P, Schoof EM, Eming SA, Smola H and Auf dem Keller U (2022). Longitudinal evaluation of biomarkers in wound fluids from venous leg ulcers and split-thickness skin graft donor site wounds treated with a protease-modulating wound dressing. Acta Derm Venereol, 102, adv00834.
- Sabino, F, Egli, FE, Savickas, S, Holstein, J, Kaspar, D, Rollmann, M, Kizhakkedathu, JN, Pohlemann, T, Smola, H and auf dem Keller U (2018). Comparative degradomics of porcine and human wound exudates unravels biomarker candidates for assessment of wound healing progression in trauma patients. J Invest Dermatol 138, 413-422.
- Seltmann, K+, *Meyer, M+, Sulcova, J, Kockmann, T, Wehkamp, U, Weidinger, S, auf dem Keller U* and Werner S* (2018). Humidity-regulated CLCA2 protects the epidermis from hyperosmotic stress. Sci Transl Med 10, eaa04650. +Joint first authors, *corresponding authors.
- Schlage P, Kockmann T, Sabino F, Kizhakkedathu JN and auf dem Keller U (2015). Matrix metalloproteinase 10 degradomics in keratinocytes and epidermal tissue identifies bioactive substrates with pleiotropic functions. Mol Cell Proteomics 14, 3234-3246.
- Sabino F+, Hermes O+, Egli, FE, Kockmann T, Schlage P, Croizat P, Kizhakkedathu JN, Smola, H and auf dem Keller U (2015). In vivo assessment of protease dynamics in cutaneous wound healing by degradomics analysis of porcine wound exudates. Mol Cell Proteomics 14, 354-370. +Joint first authors.